 Image credit: Unsplash
Image credit: Unsplash
Abstract
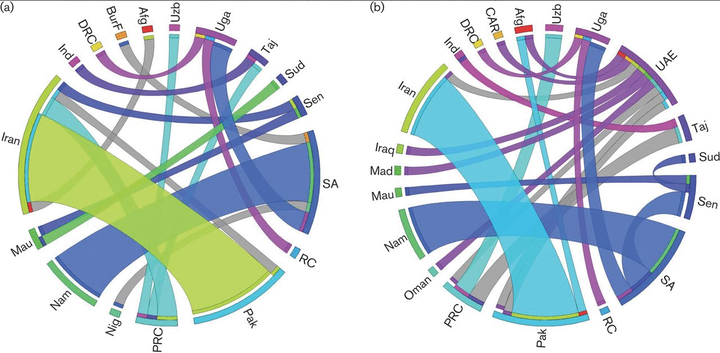
Crimean–Congo haemorrhagic fever virus (CCHFV) is a tick-borne virus with high pathogenicity to humans. CCHFV contains a three-segment [small (S), medium (M) and large (L)] genome and is prone to reassortment. Investigation of identified reassortment events can yield insight into the evolutionary history of the virus, while migration events reflect its geographical dissemination. While many studies have already considered these issues, they have investigated small numbers of isolates and lack statistical support for their findings. Here, we consider a larger set of 30 full genomes to investigate reassortment using recombination methods, as well as two sets of partial S segments comprising 393 isolates, reflecting a broader geographical range, to investigate migration events. Phylogenetic analysis revealed that the S segment showed strong geographical subdivision, but this was less apparent in the M and L segments. A total of 16 reassortment events with 22 isolates were identified with strong statistical support. Migration analysis on the partial S segments identified both long- and short-range migration events that spanned the entire geographical region in which the CCHFV has been isolated, reflecting the complex processes associated with the dissemination of the virus.