Genome comparison provides molecular insights into the phylogeny of the reassigned new genus Lysinibacillus
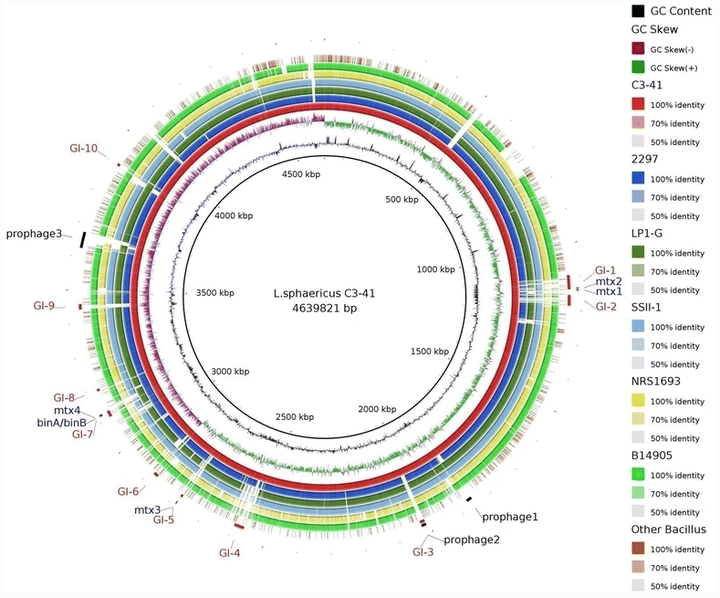 Image credit: Unsplash
Image credit: Unsplash
Abstract
Lysinibacillus sphaericus (formerly named Bacillus sphaericus) is incapable of polysaccharide utilization and some isolates produce active insecticidal proteins against mosquito larvae. Its taxonomic status was changed to the genus Lysinibacillus in 2007 with some other organisms previously regarded as members of Bacillus. However, this classification is mainly based on physiology and phenotype and there is limited genomic information to support it. In this study, four genomes of L. sphaericus were sequenced and compared with those of 24 representative strains belonging to Lysinibacillus and Bacillus. The results show that Lysinibacillus strains are phylogenetically related based on the genome sequences and composition of core genes. Comparison of gene function indicates the major difference between Lysinibacillus and the two Bacillus species is related to metabolism and cell wall/membrane biogenesis. Although L. sphaericus mosquitocidal isolates are highly conserved, other Lysinibacillus strains display a large heterogeneity. It was observed that mosquitocidal toxin genes in L. sphaericus were in close proximity to genome islands (GIs) and mobile genetic elements (MGEs). Furthermore, different copies and varying genomic location of the GIs containing binA/binB was observed amongst the different isolates. In addition, a plasmid highly similar to pBsph, but lacking the GI containing binA/binB, was found in L. sphaericus SSII-1. Our results confirm the taxonomy of the new genus Lysinibacillus at the genome level and suggest a new species for mosquito-toxic L. sphaericus. Based on our findings, we hypothesize that (1) Lysinibacillus strains evolved from a common ancestor and the mosquitocidal L. sphaericus toxin genes were acquired by horizontal gene transfer (HGT), and (2) capture and loss of plasmids occurs in the population, which plays an important role in the transmission of binA/binB.