 Photo by rawpixel on Unsplash
Photo by rawpixel on Unsplash
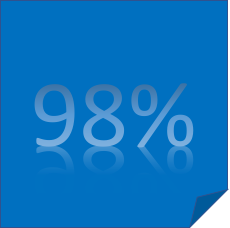
Less than 2% of the human genome consists of protein-coding genes. Initially, the remainder of the genome was assumed to be ‘junk’ DNA but it is now clear that these regions play important regulatory roles. We are interested in the regions that encode miRNA related information (e.g. transcription recognition sites, pre-miRNAs and 3’UTRs ) and how they deviate from the information in miRBase, the most commonly used miRNA reference resource. These deviations can be significant as they can impact miRNA expression studies (in the form of microArray, qPCR or Next Generation Sequencing). For example, in Next Generation Sequencing, can be in the form of miscounted miRNA reads. When combined with changes in 3’UTR targets, this can disrupt the miRNA-mRNA target events.
We are interested in investigating these effects in the context of ethnicity as reference resources rarely take this factor into account. This work is financed by a Research Council of Norway FRIPRO grant.